Harnessing the Power of GPUs to Speed Medical Imaging
Story by Helen Hill
Researchers from Northeastern University are using computers at the MGHPCC to improve the performance of a popular medical imaging tool which estimates 3D light distribution in biological tissue using GPU technology to simultaneously simulate the paths of large numbers of independent photons.
The CPU (central processing unit) has long been explained as the brain of the personal computer (PC) but over the past decade, that brain has been enhanced by another component – the GPU (graphics processing unit.)
All PCs have chips that render the display images to monitors. But not all these chips are created equal. Back in the early 2000s, integrated graphics control chips provided only basic graphics and were able to display only low resolution productivity applications like Microsoft PowerPoint, low-resolution video and basic games.
Today's generation of GPUs are in a class of their own – going far beyond basic graphics controller functions, they have become a programmable and powerful computational device in their own right.
The GPU’s advanced capabilities, which rely on parallel processing - where independent calculations are executed simultaneously - were originally used primarily for 3D game rendering. But now those capabilities have been harnessed more broadly to accelerate computational workloads in areas from astronomy and astrophysics to video and image processing for virtual and augmented reality.
Monte Carlo (MC) methods are a broad class of computational algorithms that rely on repeated random sampling to obtain numerical results. Their generality and accuracy make MC methods well-suited to simulate complex systems that may not be accessible to more direct computational techniques. A major weakness however, is their often low computational efficiency.
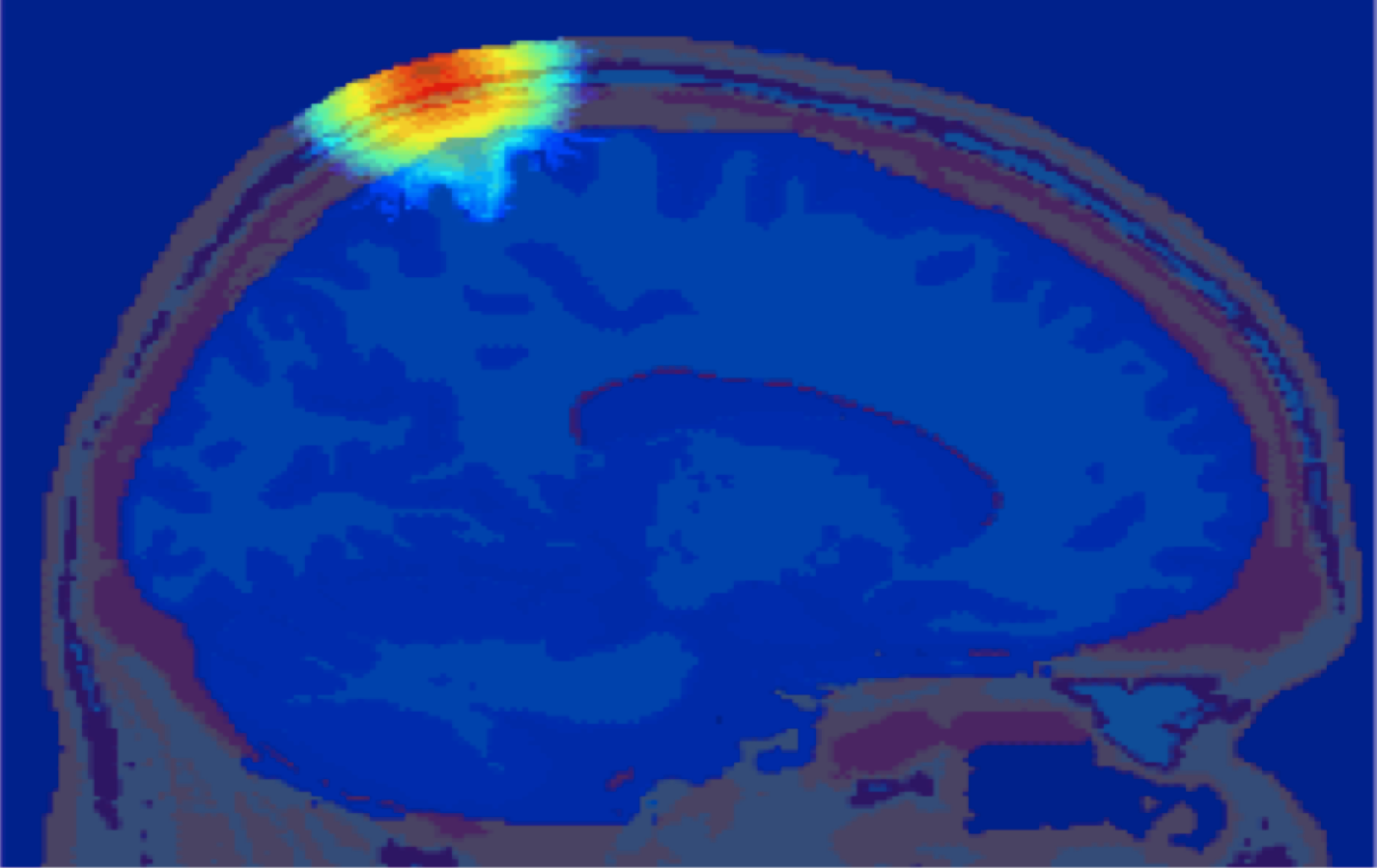
Monte Carlo simulation of photon propagation inside non-uniform tissue structures, has important applications in medical imaging, such as brain scans, but it is extremely computationally intensive.
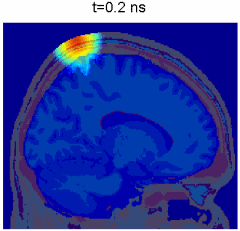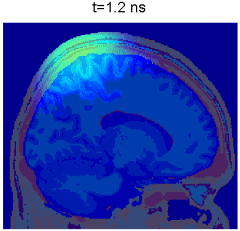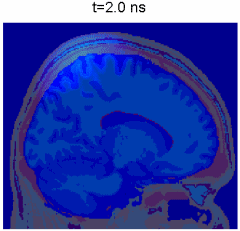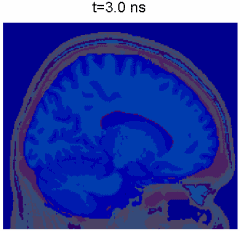
Simulation of photon transport inside human brain - animation courtesy the researchers
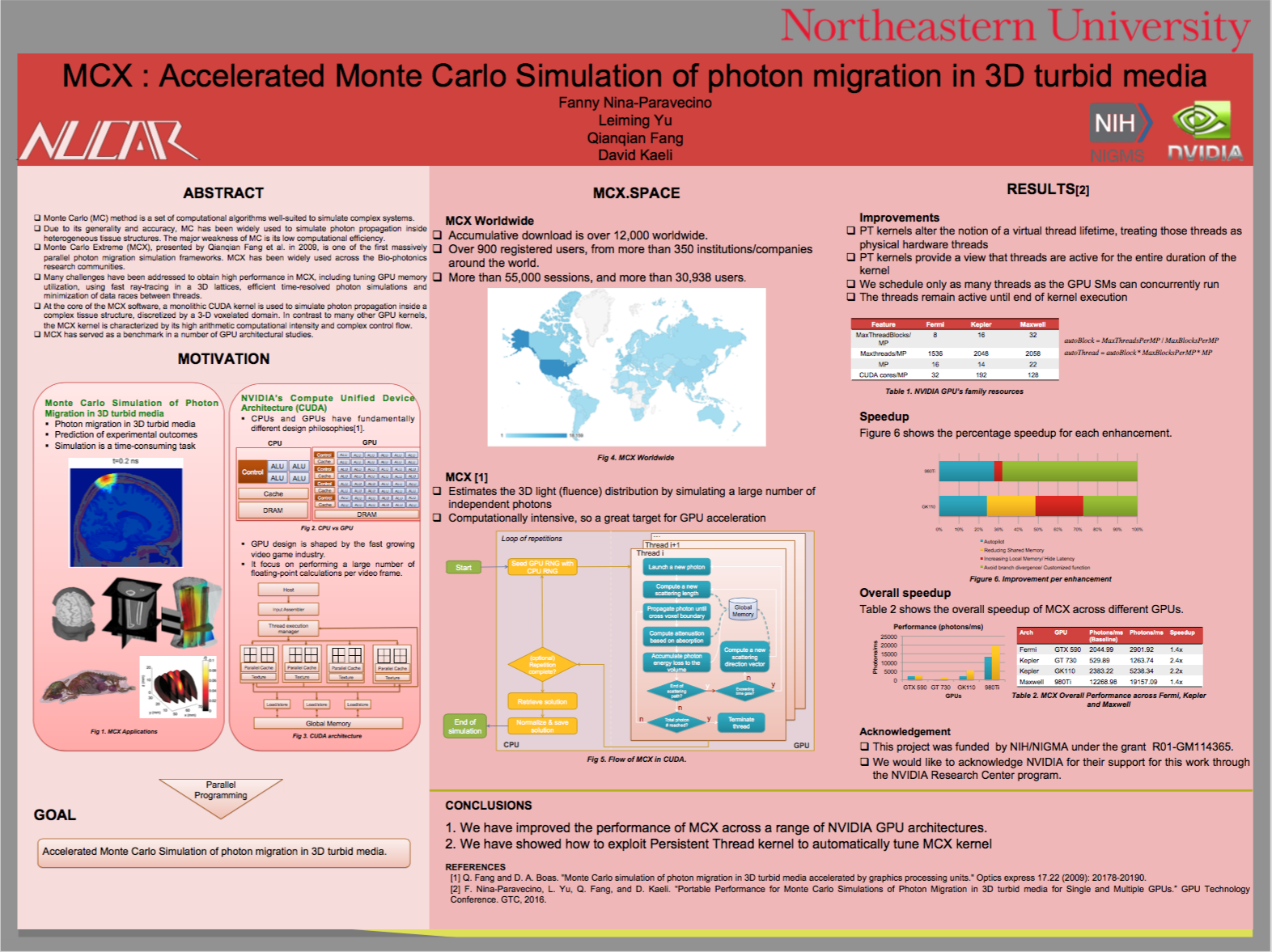
To compensate for this, Qianqian Fang, formerly a biomedical imaging researcher at the Massachusetts General Hospital, since September 2015 an Assistant Professor of Bioengineering at Northeastern University, developed Monte Carlo eXtreme (MCX for short), one of the first massively parallel photon migration simulation frameworks. Now widely used across the bio-photonics research communities, MCX has clocked up over 12000 cumulative downloads, with over 30, 000 users to-date worldwide.
Monte Carlo eXtreme, or MCX, is a Monte Carlo simulation software for time-resolved photon transport in 3D turbid media. It uses GPU based on massively parallel computing techniques and is extremely fast compared to the traditional single-threaded CPU-based simulations. Using an NVIDIA GTX 980 graphics card, the acceleration is about 300x-400x compared to a single thread on an Intel Core i7 4770k.
Many challenges have been addressed to obtain high performance in MCX, including tuning GPU memory utilization, using fast ray-tracing in 3D lattices, efficient time-resolved photon simulations, and minimization of data races between threads, however Qianqian Fang and his new colleagues at Northeastern Fanny Nina-Paravecino, Leiming Yu, and David Kaeli, continue to innovate the improvement of MCX's performance across different GPU architectures.
At HPC Day 2016, hosted by UMass Dartmouth, Fanny Nina-Paravecino, Leiming Yu, Qianqian Fang and David Kaeli, presented recent further efforts to improve the performance of MCX across a range of NVIDIA GPU architectures by exploiting "Persistent Thread kernel" to automatically tune the MCX kernel. Persistent threads kernel allows MCX to have better control of the software threads, and bring them closer to the execution lifetime of physical hardware threads in such way that it can efficiently exploit any GPU hardware using a proper grid configuration. The two main advantages of the persistent threads kernel are: a) exploiting maximal launch per Stream Multiprocessor (SM), and b) better control of the thread-block scheduling. Using this technique MCX has been improved by 20% on average across Fermi, Kepler and Maxwell architectures.
Alongside persistent threads improvement, MCX has better random generation, a new next_after, and reduced branch divergences. Overall, MCX has been improved by 100% from 2015 version.
Future work for MXC is to leverage efficiently the use of multiple GPUs, and exploit the benefit of SASSI (NVIDIA's SASS Instrumentation tool) to analyze MCX dynamic behavior.
About Fanny Nina-Paravecino:

Fanny Nina-Paravecino
Fanny Nina-Paravecino is a PhD candidate and member of the Northeastern University Computer Architecture Research Group (NUCAR), under supervision of Dr. David Kaeli.
Current research centers on high performance analysis and characterization of parallel architecture such as GPU for different algorithms including image processing, and Monte Carlo simulation of photon migration, etc. Particular areas of focus include branch divergence, nested parallelism, and concurrent kernel execution.
Publications:
Qianqian Fang and David A. Boas (2009), Monte Carlo Simulations of Photon Migration in 3D Turbid Media Accelerated by Graphics Processing Units, Optics Express 17 22 pp 20178-20190 doi: 10.1364/OE.17.020178
Fanny Nina-Paravecino, Leiming Yu, Qianqian Fang, and David Kaeli (2016), Portable Performance for Monte Carlo Simulations of Photon Migration in 3D Turbid Media for Single and Multiple GPUs, GPU Technology Conference, APRIL 4-7, 2016 | SILICON VALLEY (links require online registration)
Related Links
Northeastern University Computer Architecture Laboratory
David Kaeli
Fast photon transport simulator powered by GPU-based parallel computing Monte Carlo Extreme MCX
Qianqian Fang
NVIDIA Corporation