Vijaya B. Kolachalama, Ph.D., is an Assistant Professor at the Boston University School of Medicine. His area of expertise is in computational biomedicine and in particular machine learning and computer vision.
Activity in the Kolachalama Lab falls into two broad categories: machine learning and computer vision for precision medicine, and research into device-artery interactions, interfacial mechanics and drug delivery. In his machine learning work, Kolachalama makes extensive use of BU's Shared Computing Cluster housed at the MGHPCC.
"Artificial intelligence is poised to help deliver precision medicine, yet achieving this goal is nontrivial," says Kolachalama. "Machine learning and image processing techniques along with developments in software and hardware technologies allow us to consider questions across a range of scales," he continues. "In the radiology and digital pathology work in my lab, we leverage these tools for pattern recognition and understanding pathophysiological mechanisms, paving the way for the development of new and, we hope, more effective and accessible, diagnostic and prognostic biomedical technologies geared to a range of diseases."

Vijaya Kolachalama is an Assistant Professor of Medicine at Boston University's Medical School. In his work, he uses BU's Shared Computing Cluster, housed at the MGHPCC, in research seeking to apply machine learning and computer vision for precision medicine.
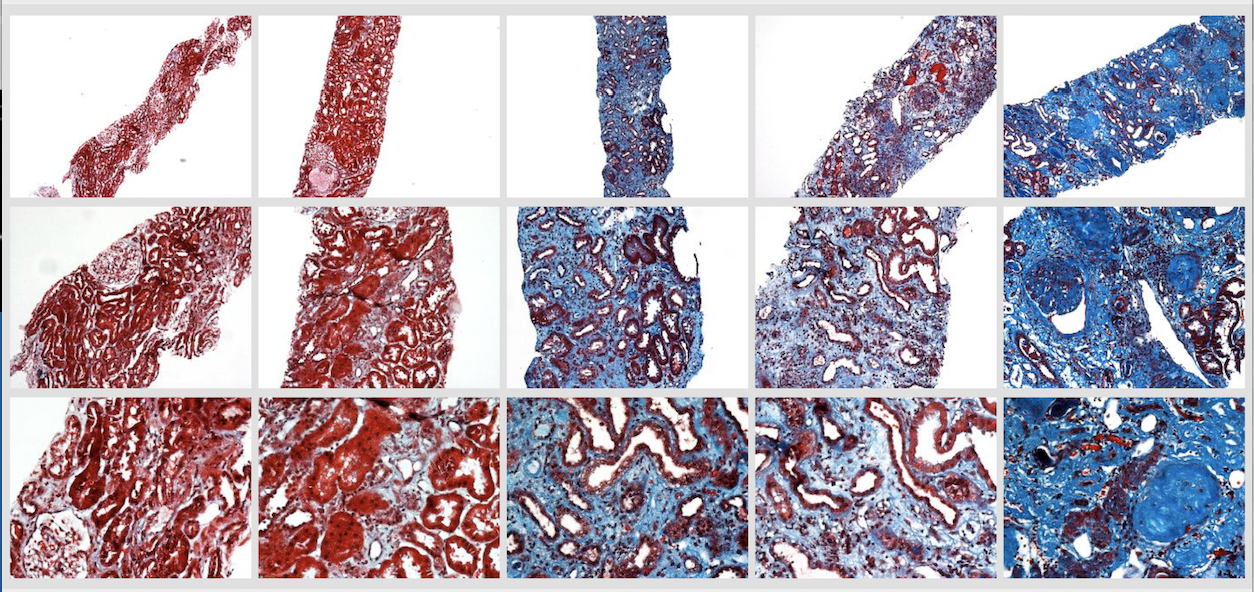
A 2018 paper about his use of deep neural networks to help in the assessment of chronic kidney disease (Kolachalama, 2018) exemplifies his lab's approach and methodologies, applying advanced machine learning techniques to systematize digital pathology.
"Chronic kidney damage is routinely assessed semiquantitatively by scoring the amount of disease seen in a renal biopsy sample," explains Kolachalama. "Although image digitization and morphometric techniques have made quantifying the extent of damage easier, the advanced machine learning tools we are developing provide a more systematic way to stratify kidney disease severity."
Speaking to BU News at the time, Kolachalama said, that "While the trained eyes of expert pathologists are able to gauge the severity of disease and detect nuances of kidney damage with remarkable accuracy, such expertise is not available in all locations, especially at a global level". Recognizing the potential of his team's model to act as a surrogate nephropathologist, especially in resource-limited settings, Kolachalama noted that “If healthcare providers around the world can have the ability to classify kidney biopsy images with the accuracy of a nephropathologist right at the point-of-care, then this can significantly impact practice."
More recently, Kolachalama has applied his machine learning techniques similarly in other areas including Alzheimer’s disease, and osteoarthritis.
"It remains difficult to characterize the source of pain in knee joints either using radiographs or magnetic resonance imaging (MRI)," he explains. "In work with Gary Chang (Chang et al, 2019), a Postdoctoral Associate in my lab, we were interested to see if using deep neural networks could distinguish knees with pain from those without it as well as to perhaps identify the structural features that are associated with knee pain."
In that study, the team constructed a convolutional Siamese network to associate MRI scans obtained on subjects from the NIH's Osteoarthritis Initiative with frequent unilateral knee pain, comparing the knee with frequent pain to the contralateral knee without pain in order to map model-predicted regions of high pain association. An expert radiologist then compared the MRI scans with the derived maps to identify the presence of abnormalities.
The radiologist's review revealed that about 86% of the cases that were predicted correctly had effusion-synovitis within the regions that were most associated with pain, suggesting deep learning can be applied to assess knee pain from MRI scans.
In the context of Alzheimer's disease, in a study, this time working with Shangran Qiu, a graduate student in the Physics Department at BU, Kolachalama and co-authors applied their machine learning tools to explore whether by combining MRI data with results from the Mini–Mental State Examination (MMSE) and logical memory tests the accuracy of diagnosing mild cognitive impairment could be enhanced (Qui et al, 2019.)
"We combined deep learning models trained on MRI slices to generate a fused MRI model using different voting techniques to predict normal cognition versus mild impairment. We then combined the fused MRI model with a second class of deep learning models trained on data obtained from NIH's National Alzheimer Coordinating Center database containing individuals with normal cognition and mild cognitive impairment," Kolachalama explains. "Our fused model did better than the individual models alone with an overall accuracy of over 90%"
Finally, in a collaboration between researchers in the Kolachalama Lab with researchers at Visterra Inc, a clinical-stage biotechnology company committed to developing innovative antibody-based therapies for the treatment of patients with kidney diseases and other hard-to-treat diseases, Kolachalama was recently involved in a study published in the journal Protein Engineering, Design & Selection (Wollacott et al, 2019), applying his machine-learning tools to quantify the “nativeness” of antibody sequences.
“Antibodies can be useful in treating, in particular, cancer and autoimmune diseases, and it has been shown that synthetic antibodies that more closely resemble their natural counterparts demonstrate improved rates of expression and stability,” explains Kolachalama. “Antibodies often undergo substantial engineering en route to the generation of a therapeutic candidate with good developability properties. Characterization of antibody libraries has shown that retaining native-like sequence improves the overall quality of the library. Using a bi-directional long short-term memory (LSTM) network model to score sequences for their similarity to naturally occurring antibodies, we were able to demonstrate our model was able to outperform other approaches at distinguishing human antibodies from those of other species.”
"None of this work would be possible without access to BU's Shared Computing Cluster and by extension the Massachusetts Green High Performance Computing Center in Holyoke where it is housed," says Kolachalama. "Our access to them is indispensable in advancing our work towards developing clinically useful digital pathology tools."
Story image: Trichrome-stained images from renal biopsy samples at different magnifications - image courtesy V. Kolachalama
Related Publications:
Kolachalama V.B., Singh P., Lin C.Q., Mun D., Belghasem M.E., Henderson J.M., Francis J.M., Salant D.J., Chitalia V.C.(2018), Association of Pathological Fibrosis with Renal Survival Using Deep Neural Networks, Kidney Int. Rep., doi: 10.1016/j.ekir.2017.11.002
Chang G.H., Felson D.T., Qiu S., Capellini T.D., Kolachalama V.B. (2019), Assessment of bilateral knee pain from MR imaging using deep neural networks, bioRxiv, doi: 10.1101/463497
Qiu, S., Chang G.H., Panagia M., Gopal D.M., Au R., Kolachalama V.B. (2018), Fusion of deep learning models of MRI scans, Mini–Mental State Examination, and logical memory test enhances diagnosis of mild cognitive impairment, Alzheimer's & Dementia: Diagnosis, Assessment & Disease Monitoring, doi: 10.1016/j.dadm.2018.08.013
Andrew M Wollacott, Chonghua Xue, Qiuyuan Qin, June Hua, Tanggis Bohnuud, Karthik Viswanathan, Vijaya B Kolachalama (2019), Quantifying the nativeness of antibody sequences using long short-term memory networks, Protein Engineering, Design and Selection, doi: 10.1093/protein/gzz031
Links
Kolachalama Lab
Boston University Shared Computing Cluster
New AI Technology Significantly Improves Human Kidney Analysis BU News