
Aquatic mammals and reptiles are striking examples of convergent evolution, but understanding how aquatic habits evolved in these groups presents paleontologists with a massive computational challenge. Caleb Gordon and his colleagues sought to address this challenge using a combination of classic paleontology methods, 3D data visualization, and phylogenetically informed machine-learning algorithms. Yale's high-performance computing resources made his new analytical approach tractable.
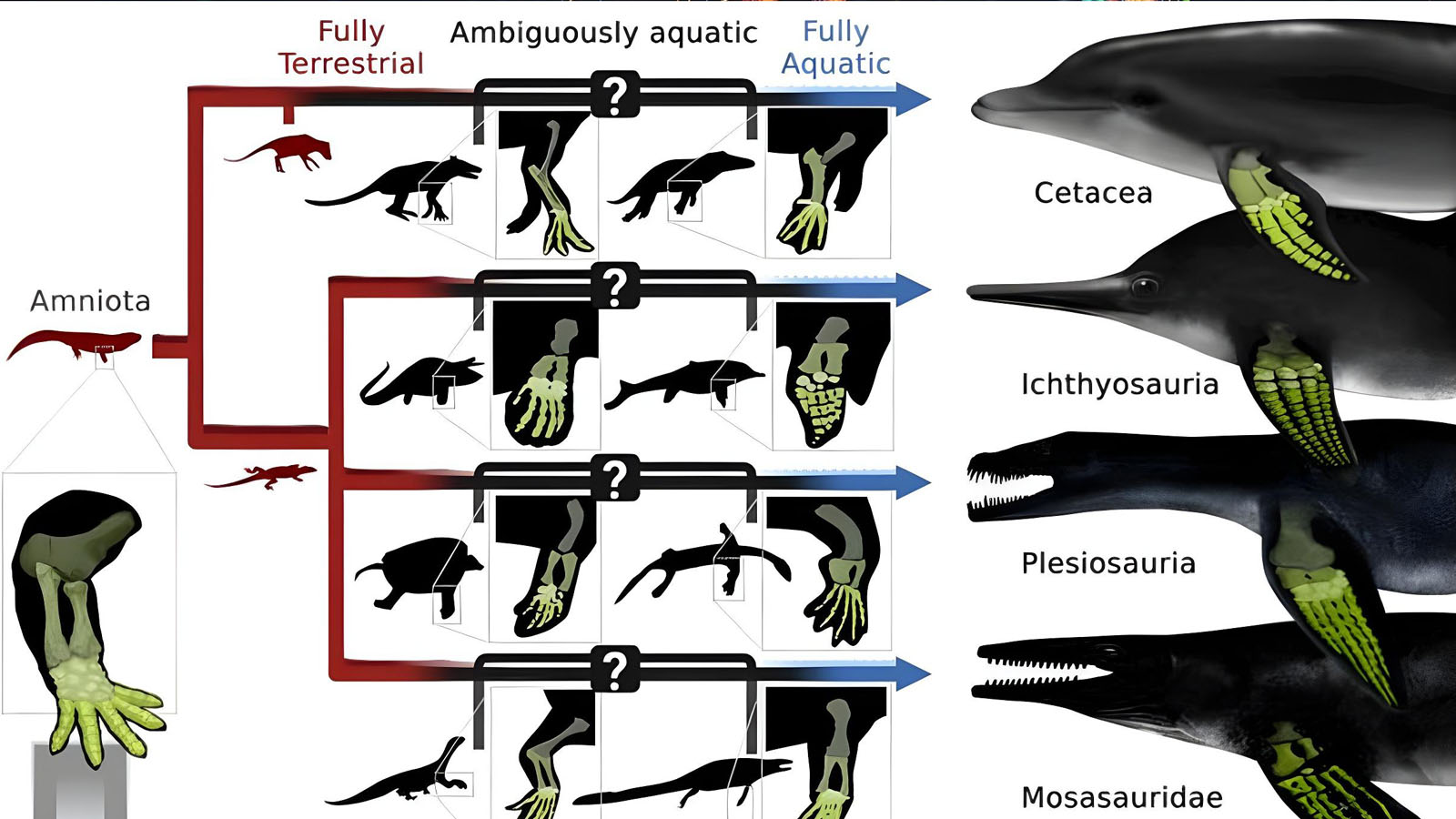
A recent paper details a machine-learning framework Gordon and co-authors developed to predict aquatic habits and soft-tissue limb features—like flippers—in extinct amniotes using fossilized bone proportions. By analyzing over 11,000 linear measurements and geometric morphometric data from 747 specimens, the team found that relative hand length is a powerful predictor of flipper presence and aquatic lifestyle, achieving over 90% accuracy across mammals and reptiles.
Their phylogenetic models revealed that while interdigital webbing cannot be reliably inferred from bones alone, flippered limbs and highly aquatic habits correlate strongly with specific forelimb proportions. Applying these models to extinct species clarified long-standing debates about the ecology of marine reptiles, showing multiple independent origins of flippers and aquatic lifestyles in groups like mosasaurs, ichthyosaurs, and thalattosaurs.
This work required extraordinary computational resources. Gordon ran multi-day job arrays involving 10,000 CPUs in parallel, consuming hundreds of thousands of CPU-hours on Yale’s Grace HPC cluster. These resources enabled complex phylogenetic regressions and simulations across millions of tree configurations -demonstrating how research computing is transforming paleobiology by revealing hidden evolutionary patterns in Earth’s history.